Set up the working environment
Overview
Teaching: 2 min
Exercises: 1 minObjectives
load packages.
set up the Maxent path.
set up working directory.
1.1 Load packages
Running Maxent in R requires several packages. Specifically, the “dismo” package, which contains maxent function that calls maxent.jar in R, the raster package, which provides functions for analyzing gridded data, the rgeos package, which provides functions for analyzing spatial data.
packages_needed <- c("raster", # for raster analysis
"dismo", # a collection of ENM/SDM tools
"rgeos","rgdal","sp", # spatial data analysis
"ENMeval", # a few new tools in ENM/SDM
"wallace", # interface for Maxent and ENMeval
"utils", # for zip & unzip files
"jsonlite" # necessary for download data from GBIF
)
pk_to_install <- packages_needed [!( packages_needed %in% rownames(installed.packages()) )]
if(length(pk_to_install)>0 ){
install.packages(pk_to_install,repos="http://cran.r-project.org")
}
library("raster")
library("dismo")
library("rgeos")
library("rgdal")
library("sp")
library("ENMeval")
#library("wallace")
if( !("rJava" %in% rownames(installed.packages())) ){
install.packages("rJava",repos="http://cran.r-project.org")
}
#If you are using a Mac machine, an additional step may be needed before loading rJava package
if(Sys.info()["sysname"] != "Windows" ){
dyn.load('/Library/Java/JavaVirtualMachines/jdk1.8.0_171.jdk/Contents/Home/jre/lib/server/libjvm.dylib')
}
library("rJava")
if(!require(devtools)){
install.packages("devtools")
}
if(!require(kuenm)){
devtools::install_github("marlonecobos/kuenm")
}
library("kuenm")
1.2 Set up the Maxent path
In order for Maxent to work properly in R, the maxent.jar file needs to be accessible by dismo package.
# download maxent.jar 3.3.3k, and place the file in the desired folder; note that, there may be a newer version of Maxent
if( !file.exists(paste0(system.file("java", package="dismo"),"/maxent.jar")) ) {
utils::download.file(url="https://raw.githubusercontent.com/mrmaxent/Maxent/master/ArchivedReleases/3.3.3k/maxent.jar",
destfile=paste0(system.file("java", package="dismo"),"/maxent.jar"),
mode="wb") ## wb for binary file, otherwise maxent.jar can not execute
}
# also note that both R and Java need to be the same bit (either 32 or 64) to be compatible to run
# to increase memory size of the JVM and prevent memory issues with Maxent.jar
# options( java.parameters = c("-Xss2560k", "-Xmx2g") )
1.3 Set up the working directory of R
By seting the working directory of R, we can directly use relative path to read/write data within this folder.
For example: an absolute path looks like this:
D:/work/project/armadillo/model/occurrence.csv
if we set the working directory as D:/work/project/armadillo, then the relative path will be:
model/occurrence.csv
setwd("~/enmClass")
#or
setwd("D:/enmClass")
# note: the folder should exist, before you set it up.
1.4 set up folders before we process data
It is recommended to setup a series of folders before you start a new project.
Here is an example from Blog of Methods Ecology & Evolution:
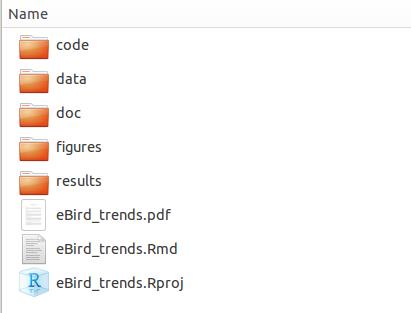 .
.
if(!file.exists("code")) dir.create("code")
if(!file.exists("data")) dir.create("data")
if(!file.exists("data/bioclim")) dir.create("data/bioclim")
if(!file.exists("data/studyarea")) dir.create("data/studyarea")
if(!file.exists("output")) dir.create("output")
Challenge 1: check if packages are setup?
Run a simple Maxent model from R terminal
Solution
# get predictor variables fnames <- list.files(path=paste(system.file(package="dismo"), '/ex', sep=''), pattern='grd', full.names=TRUE ) predictors <- stack(fnames) # file with presence points occurence <- paste(system.file(package="dismo"), '/ex/bradypus.csv', sep='') occ <- read.table(occurence, header=TRUE, sep=',')[,-1] # witholding a 20% sample for testing fold <- kfold(occ, k=5) occtest <- occ[fold == 1, ] occtrain <- occ[fold != 1, ] # fit model, biome is a categorical variable me <- maxent(predictors, occtrain, factors='biome') # see the maxent results in a browser: me
Challenge 2: Setup a working directory in R.
create a folder on your computer, and set it as the default working directory
Solution
mypath <- "~/enmClass" # or mypath <- "D:/enmClass" #create a folder dir.create(mypath) #set the working directory as mypath setwd(mypath) # read the working directory, the output should be the same as the path you set above getwd(mypath)
Challenge 3: create a folder names “temp”
Solution
dir.create("temp")