Raster data
Overview
Teaching: 10 min
Exercises: 10 minObjectives
read/write raster data
raster metadata
raster resample
raster reclassification
raster calculation
raster correlation
3.1 Load libraries and set up folders
library("raster")
require(utils)
if(!file.exists("data")) dir.create("data")
if(!file.exists("data/bioclim")) dir.create("data/bioclim")
if(!file.exists("data/studyarea")) dir.create("data/studyarea")
if(!file.exists("output")) dir.create("output")
3.2 Download data
In our example, we use bioclimatic variables (downloaded from worldclim.org) as input environmental layers.
We use download.file() to directly download worldclim data, and used unzip() to unzip the zipped file
# download climate data from worldclim.org
if( !file.exists( paste0("data/bioclim/bio_10m_bil.zip") )){
utils::download.file(url="http://biogeo.ucdavis.edu/data/climate/worldclim/1_4/grid/cur/bio_10m_bil.zip",
destfile="data/bioclim/bio_10m_bil.zip" )
utils::unzip("data/bioclim/bio_10m_bil.zip",exdir="data/bioclim")
}
3.3 Read/Write raster files
read one raster file at one time
test1 <- raster("data/bioclim/bio1.bil")
dir.create("temp")
writeRaster(test1,"temp/bio1.bil",overwrite=TRUE)
writeRaster(test1,"temp/bio1.tiff",overwrite=TRUE)
read 19 layers at one time
# search files with .bil file extension
clim_list <- list.files("data/bioclim/",pattern=".bil$",full.names = T)
# stacking the bioclim variables to process them at one go
clim <- raster::stack(clim_list)
You can write many different type of rasters:
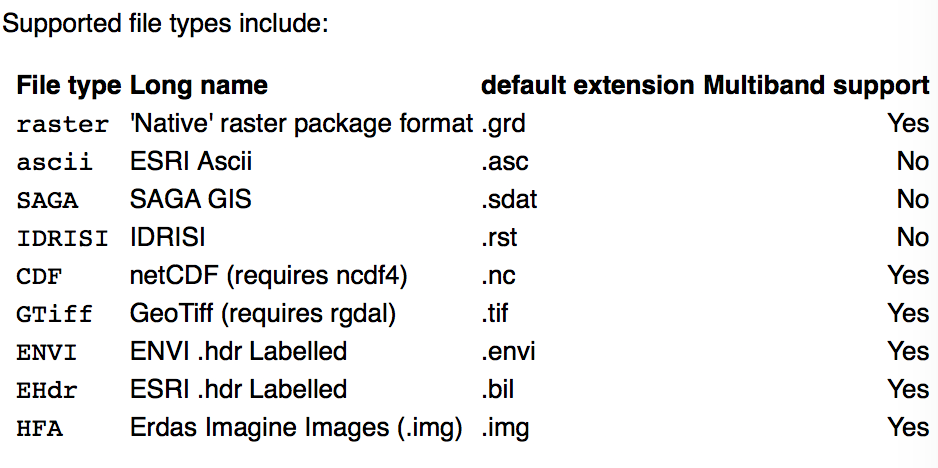 .
.
3.4 Basic information of a raster
bio1 <- raster("data/bioclim/bio1.bil")
bio1
class : RasterLayer
dimensions : 900, 2160, 1944000 (nrow, ncol, ncell)
resolution : 0.1666667, 0.1666667 (x, y)
extent : -180, 180, -60, 90 (xmin, xmax, ymin, ymax)
coord. ref. : +proj=longlat +datum=WGS84 +no_defs +ellps=WGS84 +towgs84=0,0,0
data source : /Users/lablap/lesson_mac/_episodes_rmd/data/bioclim/bio1.bil
names : bio1
values : -269, 314 (min, max)
read number of rows
nrow(bio1)
[1] 900
read number of columns
ncol(bio1)
[1] 2160
read extent
extent(bio1)
class : Extent
xmin : -180
xmax : 180
ymin : -60
ymax : 90
read CRS (coordinate reference system)
crs(bio1)
CRS arguments:
+proj=longlat +datum=WGS84 +no_defs +ellps=WGS84 +towgs84=0,0,0
read resolution
res(bio1)
[1] 0.1666667 0.1666667
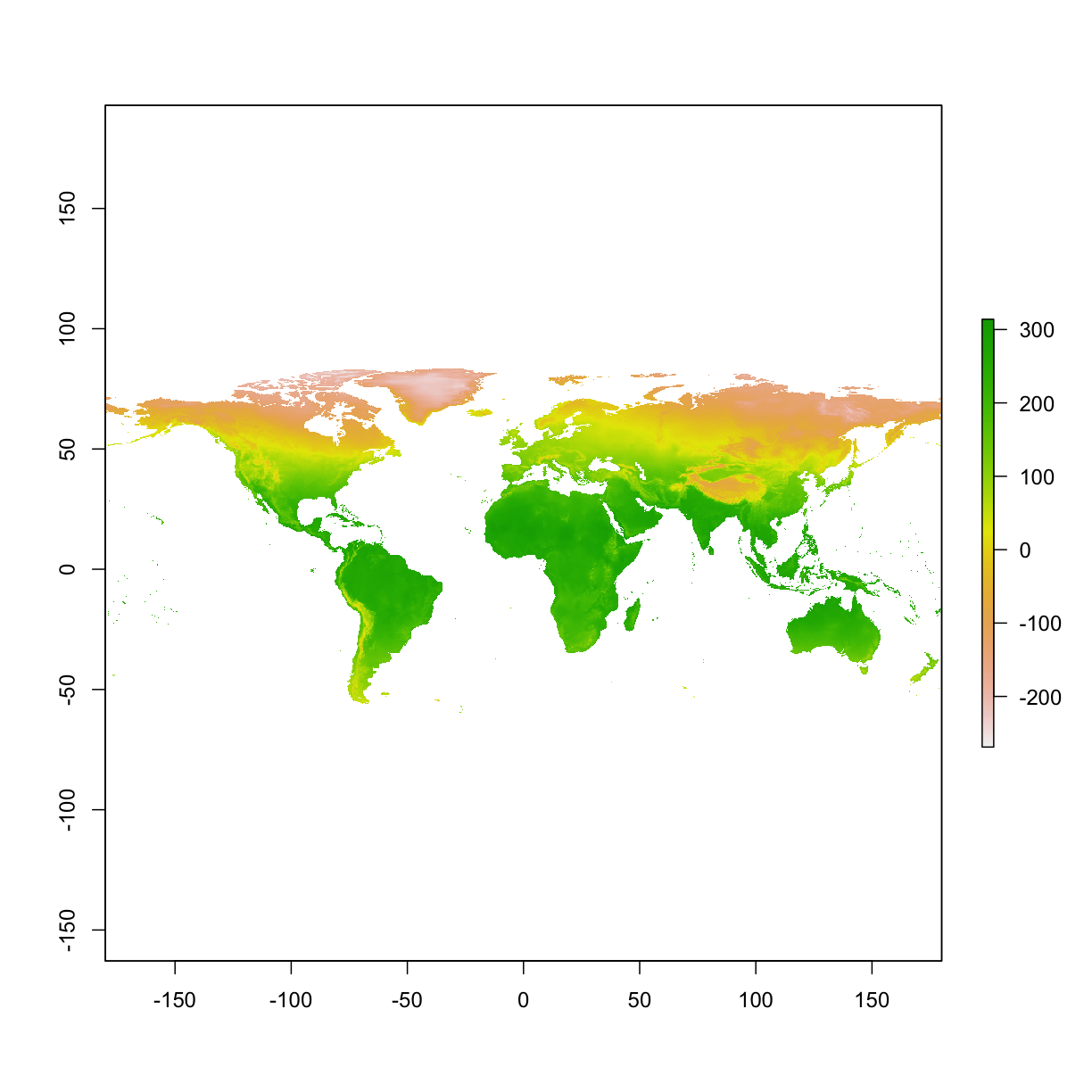
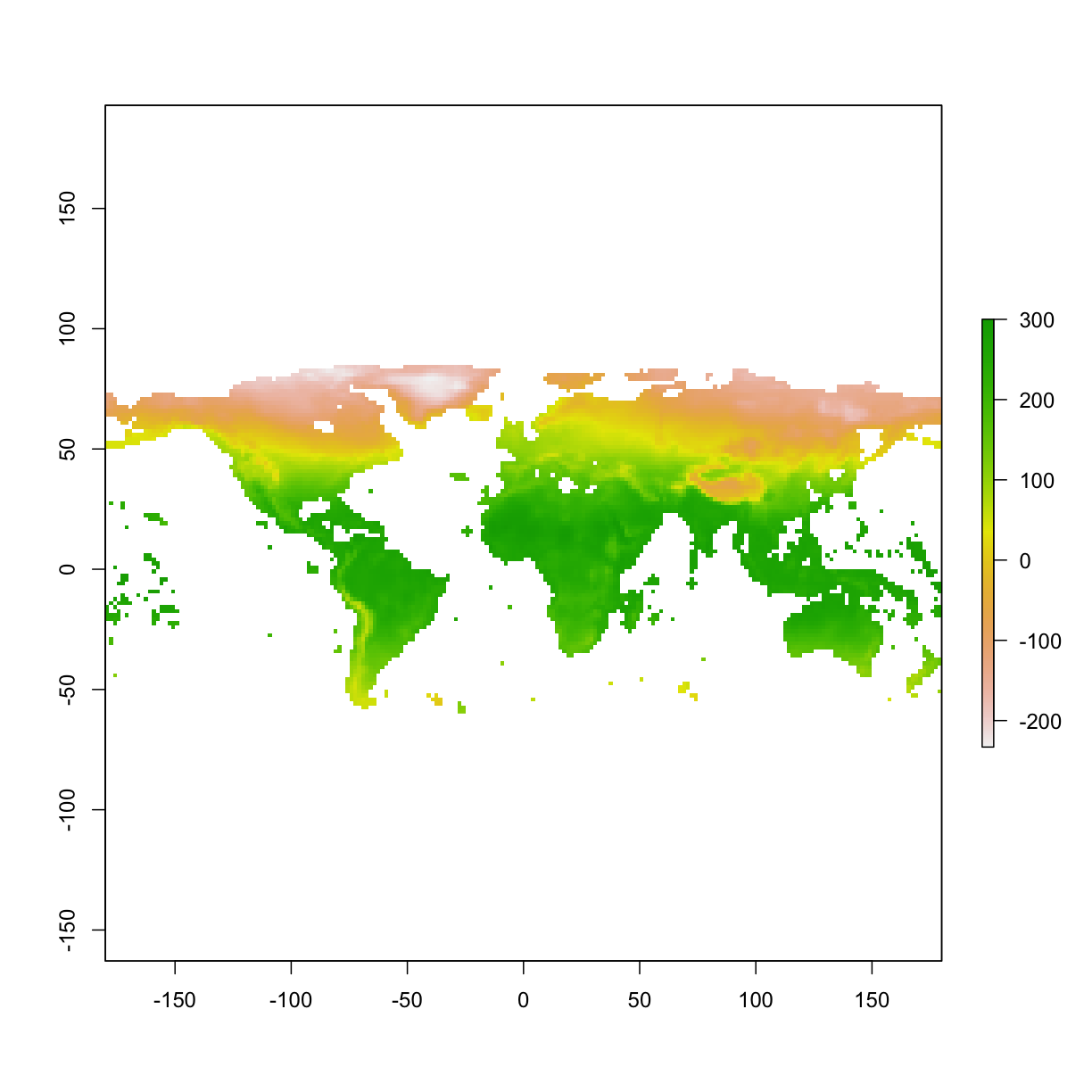
3.5 Resample raster
We want the new layer to be 10 times coarser at each axis (i.e., 100 times coarser). In essence, we are resampling the resolution from 10 min to 100 min.
bio1 <- raster("data/bioclim/bio1.bil")
# define new resolution
newRaster <- raster( nrow= nrow(bio1)/10 , ncol= ncol(bio1)/10 )
# define the extent of the new coarser resolution raster
extent(newRaster) <- extent(bio1)
# fill the new layer with new values
newRaster <- resample(x=bio1,y=newRaster,method='bilinear')
# when viewing the new layer, we see that it appears coarser
plot(bio1)
plot(newRaster)
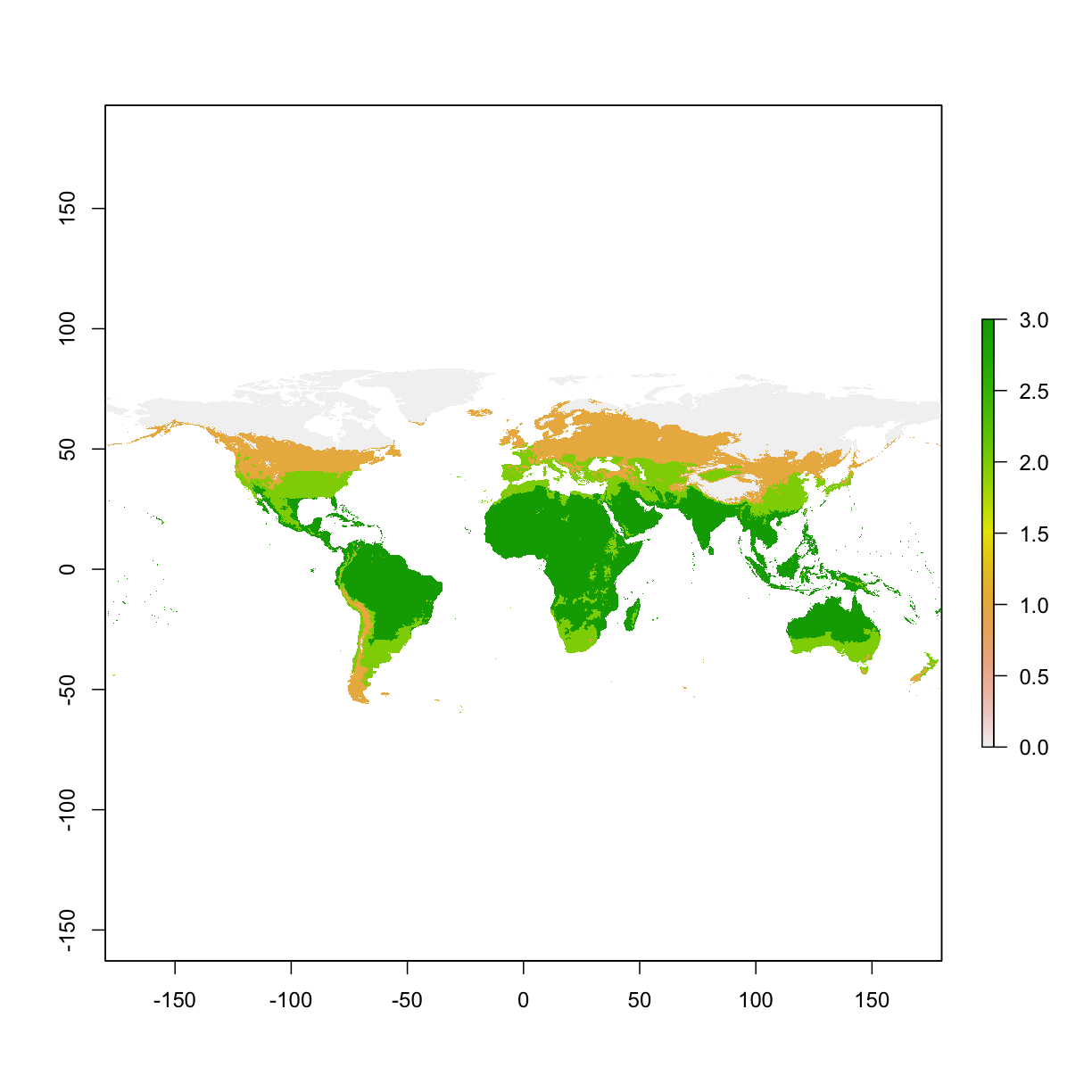
3.6 Reclassify raster layer
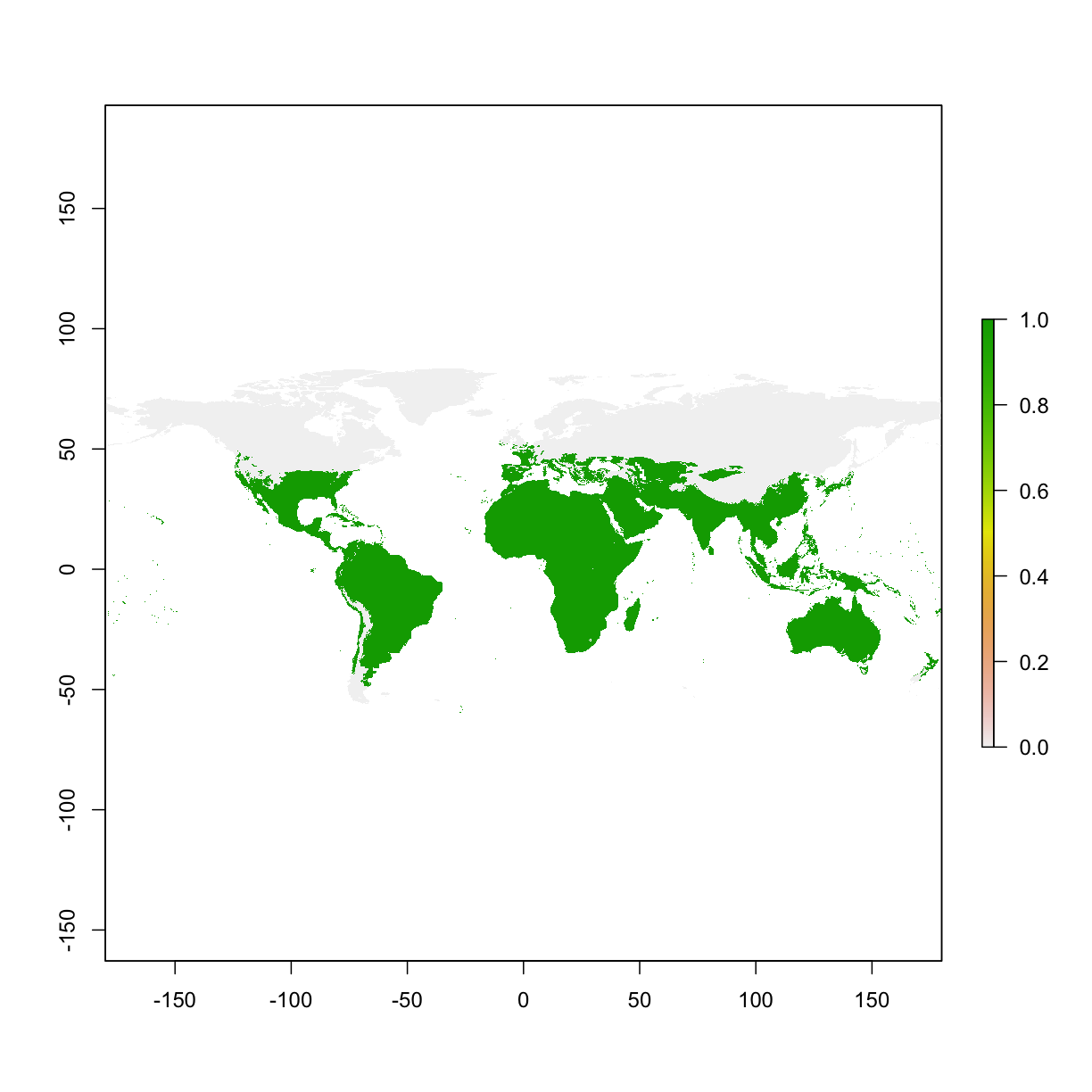
Reclassify as a binary layer: areas that are higher than 100 va. areas that are lower than 100
myLayer<- raster("data/bioclim/bio1.bil")
binaryMap <- myLayer>= 100
plot(binaryMap)
Use multiple threholds to reclassify
# values smaller than 0 becomes 0;
# values between 0 and 100 becomes 1;
# values between 100 and 200 becomes 2;
# values larger than 200 becomes 3;
myMethod <- c(-Inf, 0, 0, # from, to, new value
0, 100, 1,
100, 200, 2,
200, Inf, 3)
myLayer_classified <- reclassify(myLayer,rcl= myMethod)
plot(myLayer_classified)
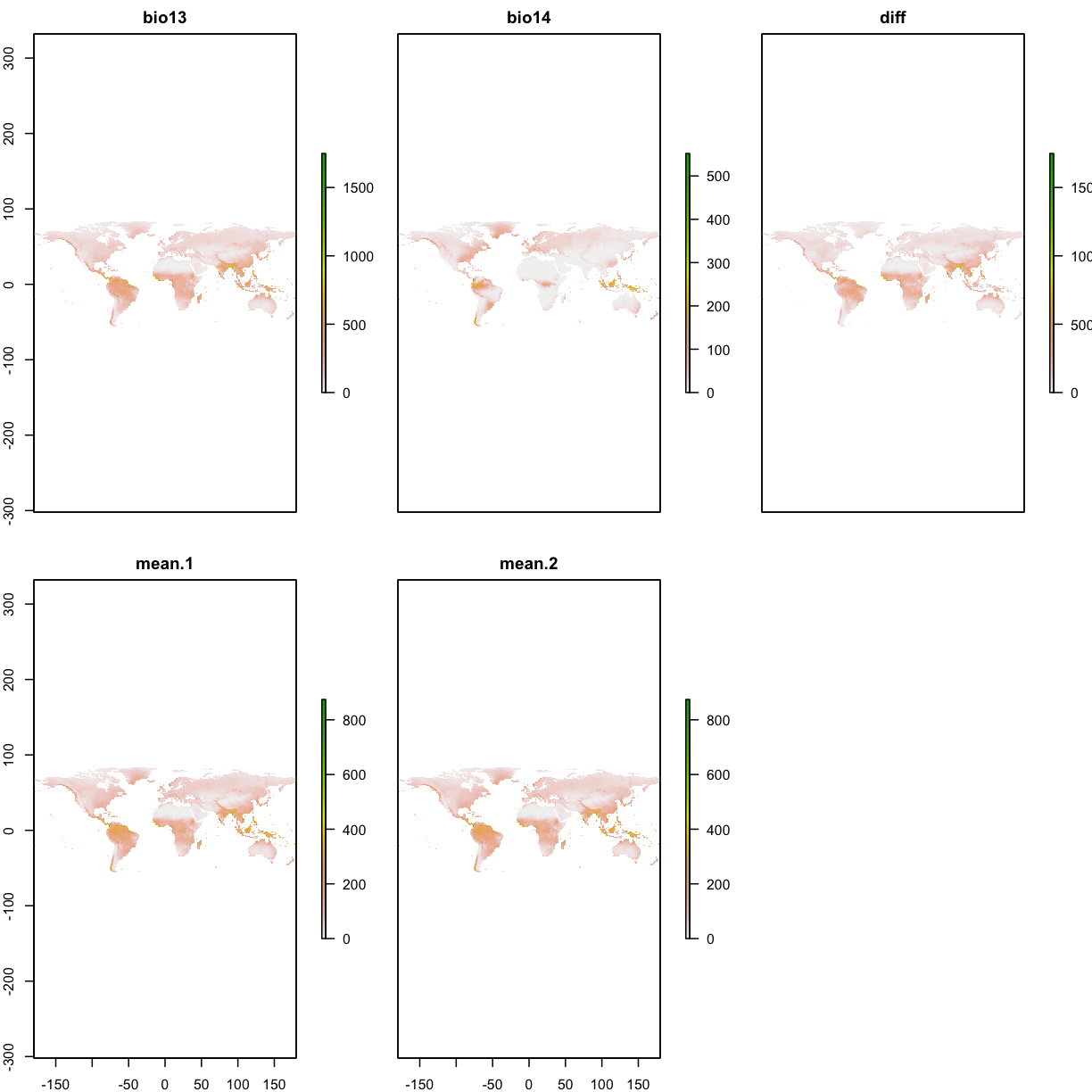
3.7 Raster calculation
wet <- raster("data/bioclim/bio13.bil") # precipitation of wettest month
dry <- raster("data/bioclim/bio14.bil") # precipitation of driest month
# To calculate difference between these two rasters
diff <- wet - dry
names(diff) <- "diff"
# To calculate the mean between the dry and wet rasters
twoLayers <- stack(wet,dry)
meanPPT1 <- calc(twoLayers,fun=mean)
names(meanPPT1) <- "mean"
# the following code gives the same results
meanPPT2 <- (wet + dry)/2
names(meanPPT2) <- "mean"
layers_to_plot <- stack(wet, dry, diff, meanPPT1,meanPPT2)
plot(layers_to_plot)
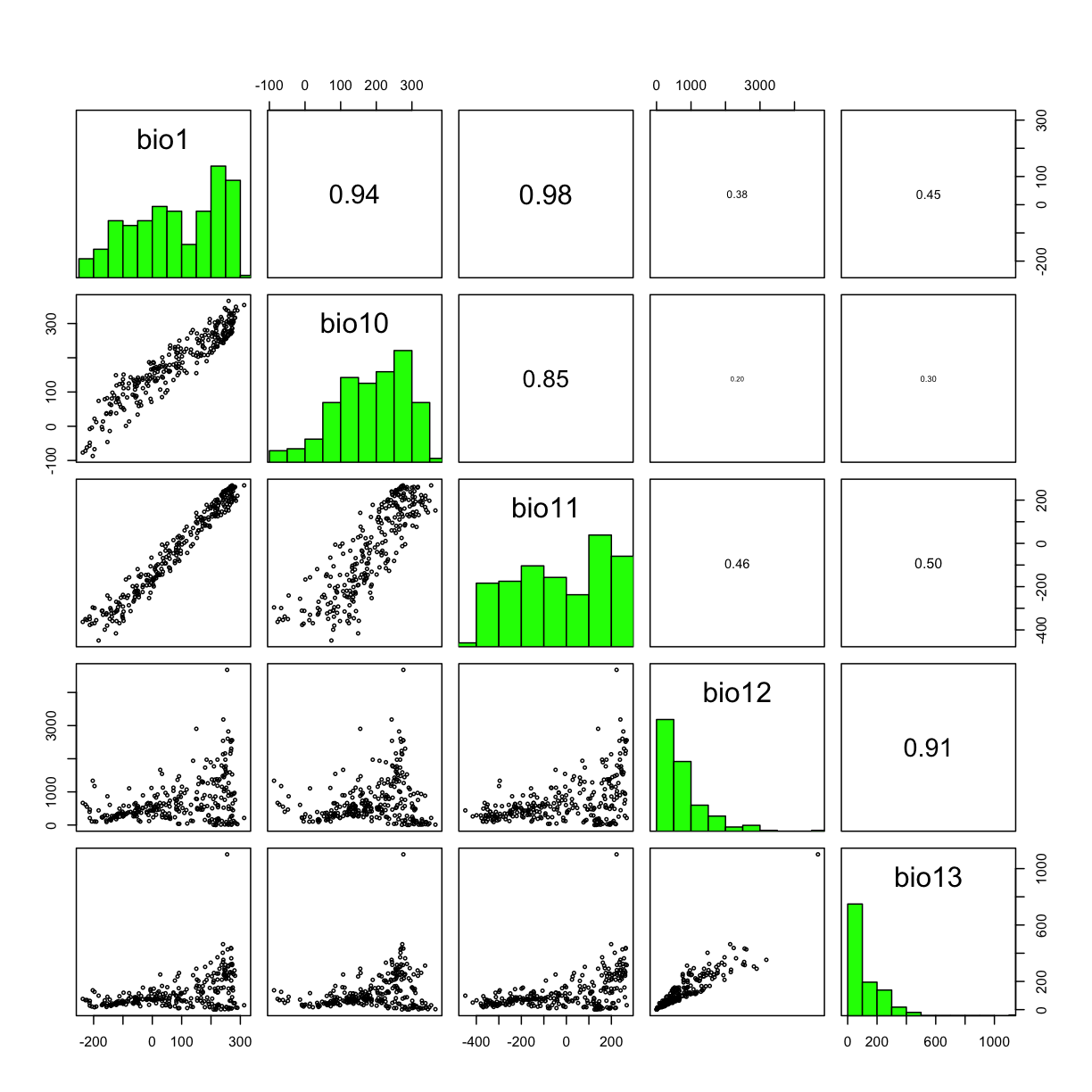
3.8 Correlations between layers
# search files with *.bil* file extension
clim_list <- list.files("data/bioclim/",pattern=".bil$",full.names = T)
# stacking the bioclim variables to process them at one go
clim <- raster::stack(clim_list)
# select the first 5 layers
clim_subset <- clim[[1:5]]
# to run correlations between different layers
raster::pairs(clim_subset,maxpixels=1000) # the default is maxpixels=100000
Challenge: download and process worldclim data
–download worldclim_V1.4 10m resolution bioclimatic layers
–read BIO5 (Max Temperature of Warmest Month) & BIO6 (Min Temperature of Coldest Month)
–calculate thedifferencebetween the two layers
–save thedifferenceasdifference.tiffin foldertempSolution
dir.create("data") dir.create("data/bioclim") # download utils::download.file(url="http://biogeo.ucdavis.edu/data/climate/worldclim/1_4/grid/cur/bio_10m_bil.zip", destfile="data/bioclim/bio_10m_bil.zip" ) utils::unzip("data/bioclim/bio_10m_bil.zip",exdir="data/bioclim") # load rasters library(raster) bio5 <- raster("data/bioclim/bio5.bil") bio6 <- raster("data/bioclim/bio6.bil") # calculate the differences difference <- bio5 - bio6 # save the raster file dir.create("temp") writeRaster(difference,"temp/difference.tiff")