Spatial analysis
Overview
Teaching: 15 min
Exercises: 10 minObjectives
plot occ & raster together
extract environmental conditions of occ raster layers
crop raster layers
draw background samples
randomly separate samples
4.0 Prepare occ & raster
library("raster")
if(!file.exists("data/occ_raw.rdata")){
occ_raw <- gbif(genus="Dasypus",species="novemcinctus",download=TRUE)
save(occ_raw,file = "data/occ_raw.rdata")
}else{
load("data/occ_raw.rdata")
}
occ_clean <- subset(occ_raw,(!is.na(lat))&(!is.na(lon)))
occ_unique <- occ_clean[!duplicated( occ_clean[c("lat","lon")] ),]
occ_unique_specimen <- subset(occ_unique, basisOfRecord=="PRESERVED_SPECIMEN")
occ_final <- subset(occ_unique_specimen, year>=1950 & year <=2000)
coordinates(occ_final) <- ~ lon + lat
myCRS1 <- CRS("+init=epsg:4326") # WGS 84
crs(occ_final) <- myCRS1
if( !file.exists( paste0("data/bioclim/bio_10m_bil.zip") )){
utils::download.file(url="http://biogeo.ucdavis.edu/data/climate/worldclim/1_4/grid/cur/bio_10m_bil.zip",
destfile="data/bioclim/bio_10m_bil.zip" )
utils::unzip("data/bioclim/bio_10m_bil.zip",exdir="data/bioclim")
}
bio1 <- raster("data/bioclim/bio1.bil")
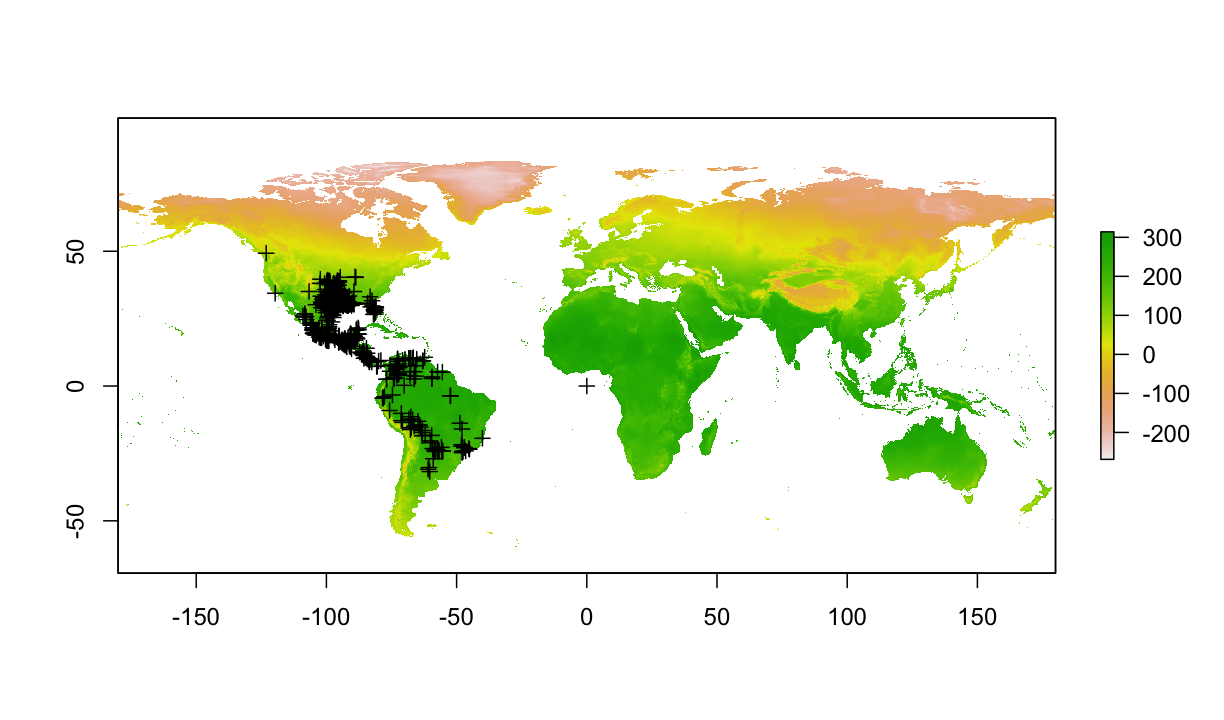
4.1 Plot occ & raster together
plot(bio1)
plot(occ_final,add=T)
4.2 Extract environmental conditions of occ raster layers
use extract() function to extract environmental conditions of occurrences
# load bioclimatic layers
clim_list <- list.files("data/bioclim/",pattern=".bil$",full.names = T)
clim <- raster::stack(clim_list)
conditions_occ <- extract(clim,occ_final)
head(conditions_occ)
bio1 bio10 bio11 bio12 bio13 bio14 bio15 bio16 bio17 bio18 bio19 bio2
[1,] 251 261 242 3170 428 67 51 1221 240 382 946 95
[2,] 262 280 242 944 263 2 112 647 13 479 35 115
[3,] 184 274 87 537 82 19 45 197 70 156 70 148
[4,] 258 276 236 866 218 2 108 592 8 485 28 133
[5,] 257 273 235 3058 517 61 67 1402 205 677 321 112
[6,] 269 286 248 993 262 1 112 594 7 305 7 102
bio3 bio4 bio5 bio6 bio7 bio8 bio9
[1,] 76 723 318 194 124 247 258
[2,] 72 1554 330 171 159 279 243
[3,] 42 7262 351 1 350 235 96
[4,] 72 1592 339 155 184 272 242
[5,] 70 1458 337 177 160 264 257
[6,] 64 1533 348 191 157 279 255
remove occurrences that have NA conditions
bad_records <- is.na( conditions_occ[,1] )
table(bad_records)
bad_records
FALSE TRUE
607 3
look at the bad records, and remove them
conditions_occ[bad_records,]
bio1 bio10 bio11 bio12 bio13 bio14 bio15 bio16 bio17 bio18 bio19 bio2
[1,] NA NA NA NA NA NA NA NA NA NA NA NA
[2,] NA NA NA NA NA NA NA NA NA NA NA NA
[3,] NA NA NA NA NA NA NA NA NA NA NA NA
bio3 bio4 bio5 bio6 bio7 bio8 bio9
[1,] NA NA NA NA NA NA NA
[2,] NA NA NA NA NA NA NA
[3,] NA NA NA NA NA NA NA
occ_final <- occ_final[!bad_records,]
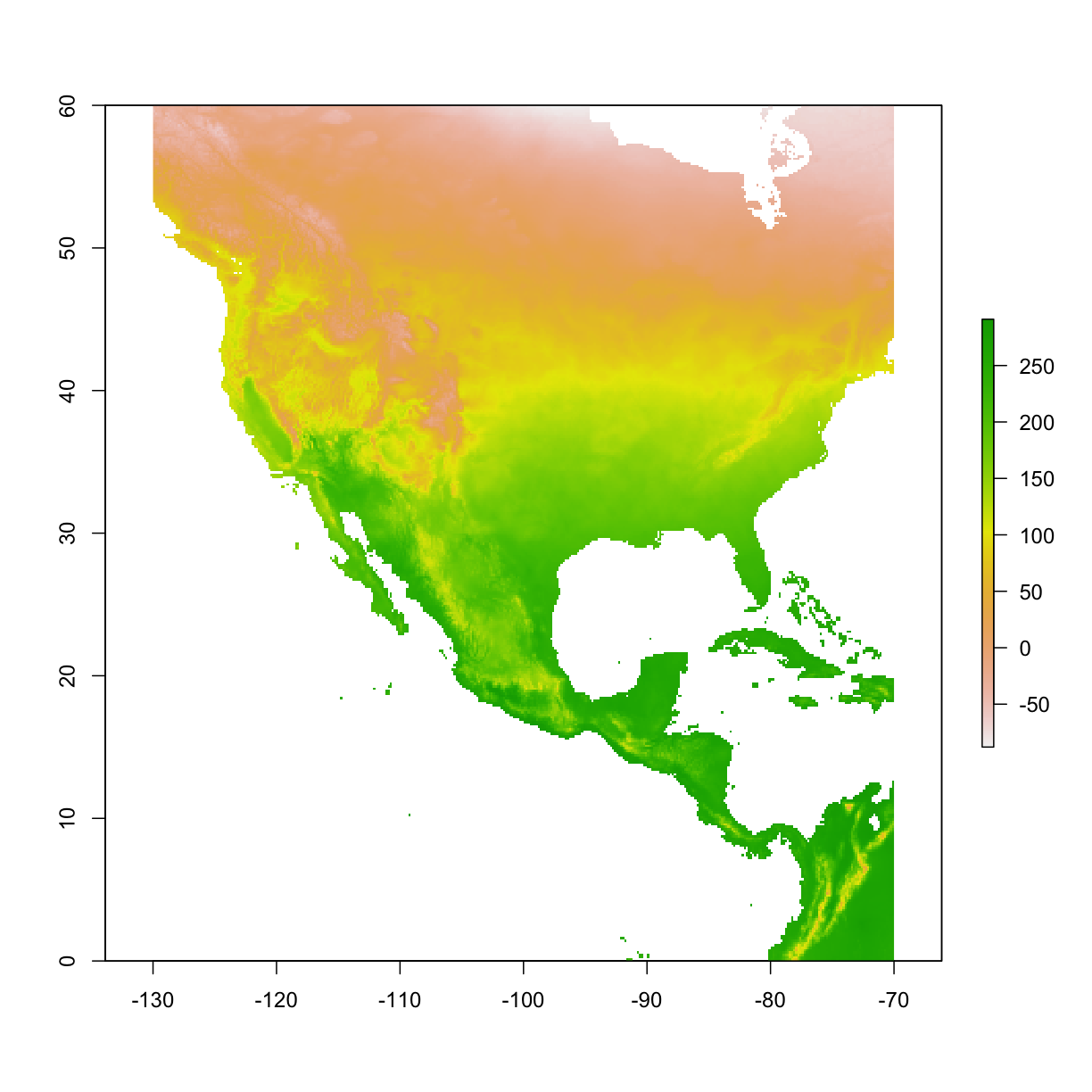
4.3 Crop raster layers
use crop() function to crop a raster layer
bio1 <- raster("data/bioclim/bio1.bil")
mybox <- extent(-130, -70, 0, 60)
#min x, max x, min y, max y
bio1_crop <- crop(bio1, mybox)
plot(bio1_crop)
 use
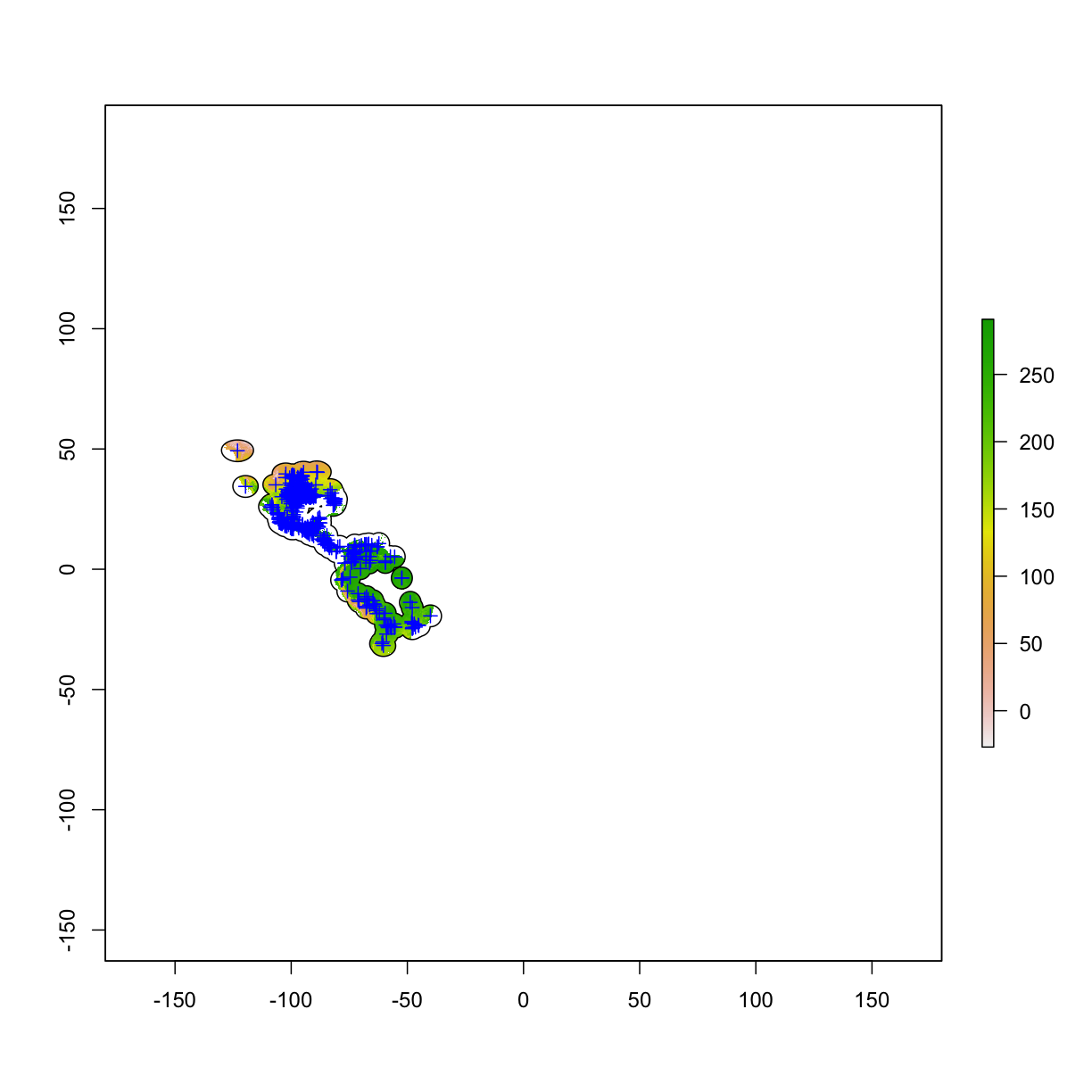
use mask() function to cut a raster with fine boundry
bio1 <- raster("data/bioclim/bio1.bil")
occ_buffer <- buffer(occ_final,width=5*10^5) #unit is meter
Loading required namespace: rgeos
bio1_mask <- mask(bio1, occ_buffer)
plot(bio1_mask)
plot(occ_buffer,add=T)
plot(occ_final,add=T,col="blue")
 this can also be applied to multiple rasters
this can also be applied to multiple rasters
# load bioclimatic layers
clim_list <- list.files("data/bioclim/",pattern=".bil$",full.names = T)
clim <- raster::stack(clim_list)
occ_buffer <- buffer(occ_final,width=5*10^5) #unit is meter
clim_mask <- mask(clim, occ_buffer)
4.4 Draw background samples
use sampleRandom() function to select background points from the new buffered study area; the number provided in the size.
use set.seed() before random sampling to guarantee the same random sample
set.seed(1)
bg <- sampleRandom(x=clim_mask,
size=10000,
na.rm=T, #removes the 'Not Applicable' points
sp=T) # return spatial points
head(bg)
bio1 bio10 bio11 bio12 bio13 bio14 bio15 bio16 bio17 bio18 bio19 bio2
1 262 270 248 1427 244 11 69 672 52 333 87 122
2 257 277 226 1118 187 25 58 495 82 476 115 113
3 249 252 245 2048 332 15 68 946 57 468 57 129
4 241 280 197 1198 147 28 39 423 126 401 134 125
5 104 106 99 1006 124 39 31 338 147 332 147 103
6 210 229 184 1415 319 7 87 793 26 575 44 120
bio3 bio4 bio5 bio6 bio7 bio8 bio9
1 70 906 347 175 172 266 251
2 66 2155 336 166 170 277 228
3 77 314 332 165 167 247 245
4 58 3291 348 133 215 274 208
5 90 281 161 47 114 105 99
6 65 1787 291 109 182 225 189
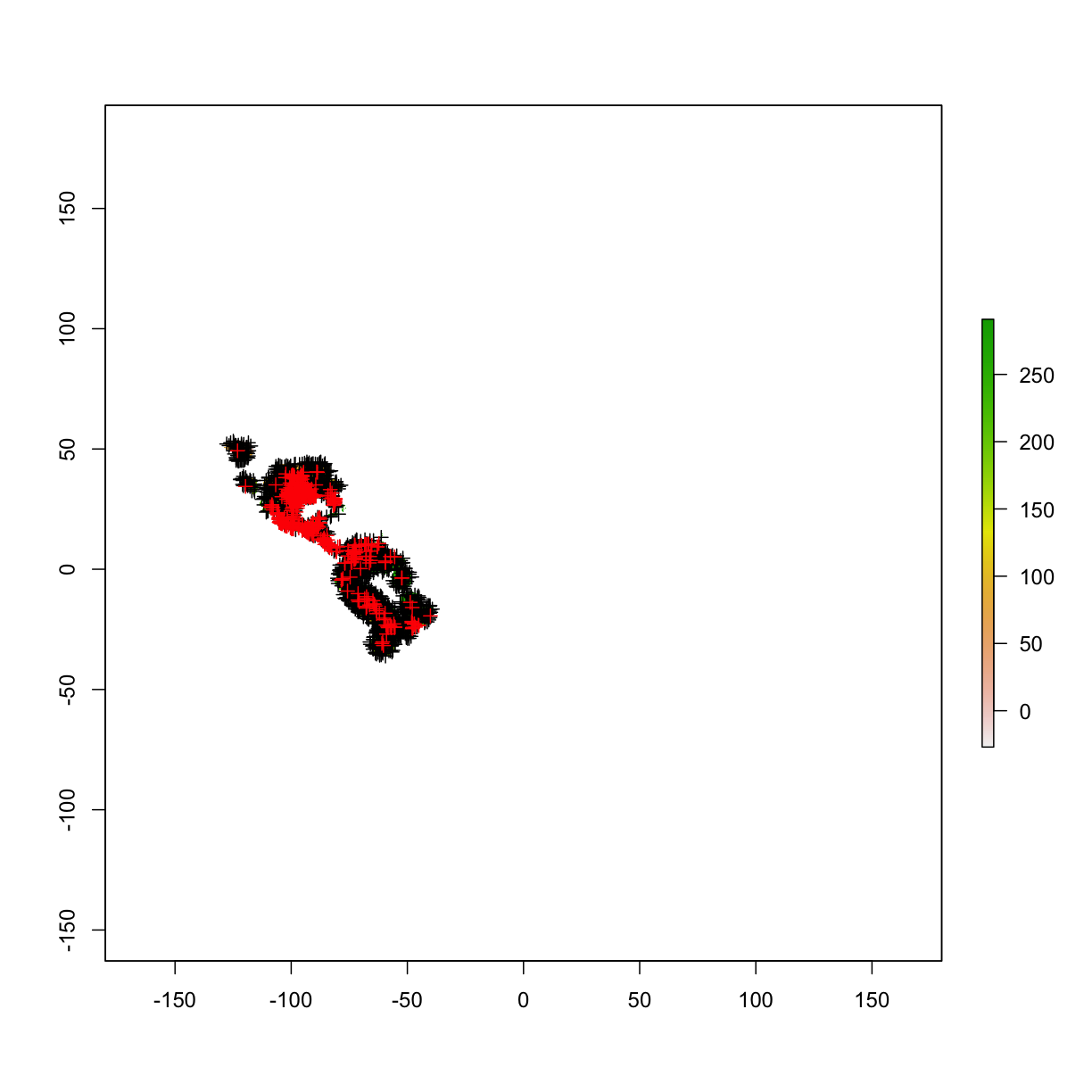
plot(clim_mask[[1]])
# add the background points to the plotted raster
plot(bg,add=T,col="black")
# add the occurrence data to the plotted raster
plot(occ_final,add=T,col="red")
4.5 Split occurrence data into training & testing
We randomly selected 50% of the occurrence data for model training and used the remaining 50% for model testing. To make our experiment reproducible (i.e., select the same set of points), we used a static seed via the set.seed() function.
# get the same random sample for training and testing
set.seed(1)
# randomly select 50% for training
selected <- sample( 1:nrow(occ_final), nrow(occ_final)*0.5)
occ_train <- occ_final[selected,] # this is the selection to be used for model training
occ_test <- occ_final[-selected,] # this is the opposite of the selection which will be used for model testing
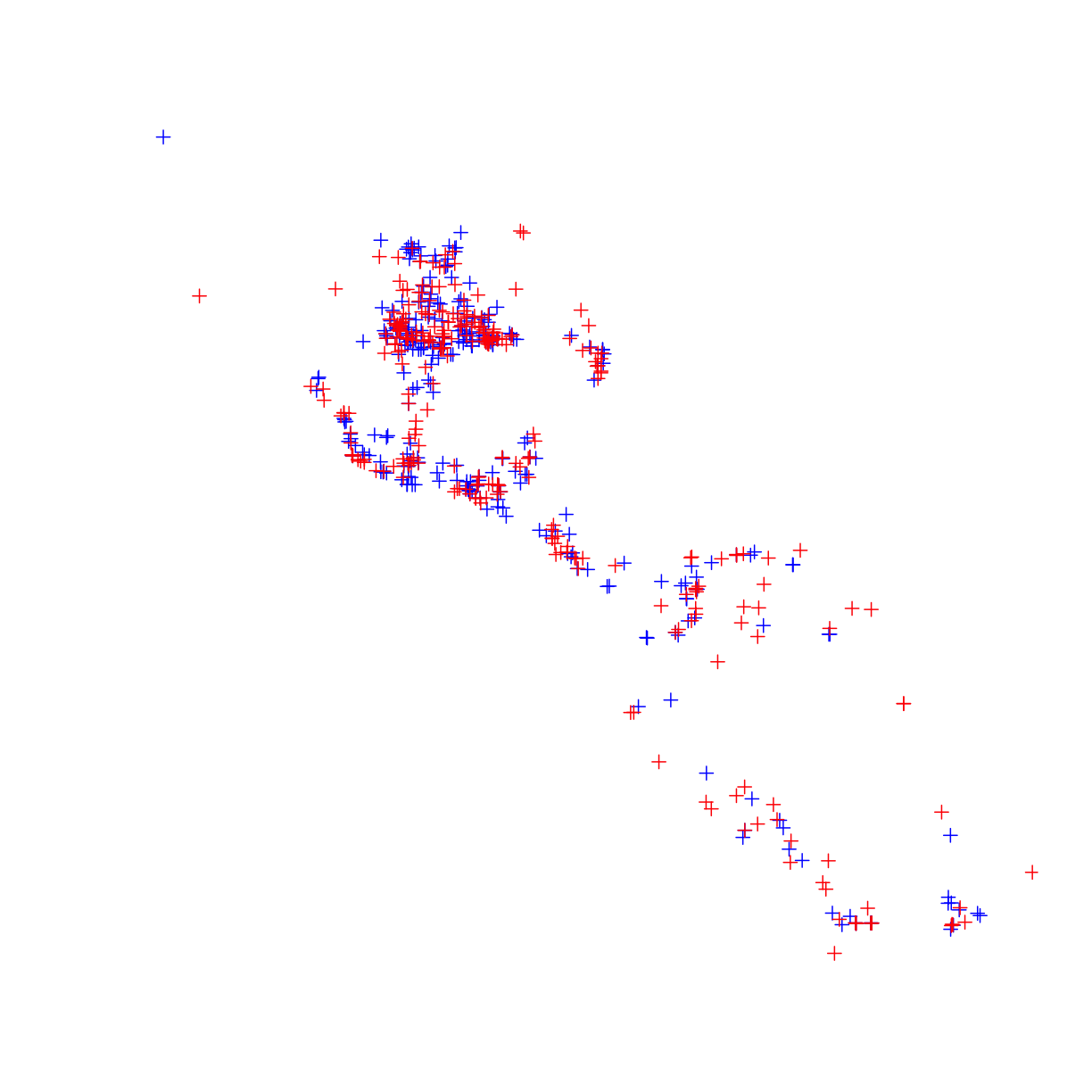
plot(occ_train,col="blue")
plot(occ_test,col="red",add=T)
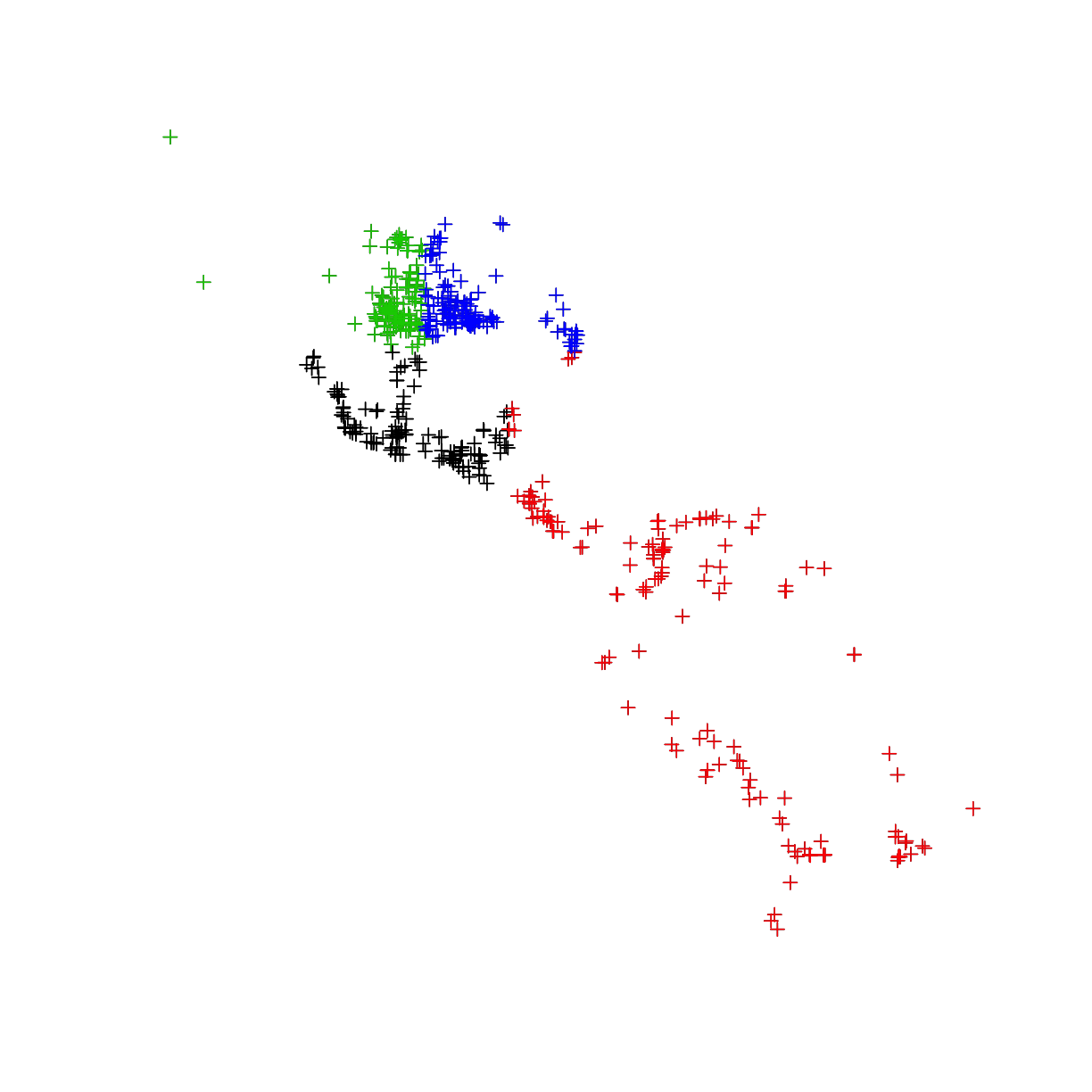
an alternative method, block cut, distributes the training and testing more equally across the study exent. This function is part of the ENMeval package.
library(ENMeval)
cut_block <- ENMeval::get.block(occ=as.data.frame(occ_final@coords),
bg.coords=as.data.frame(bg@coords))
occ_final@data$cut_block <- cut_block$occ.grp
bg@data$cut_block <- cut_block$bg.grp
plot(occ_final)
plot(subset(occ_final,cut_block==1),col=1,add=T)
plot(subset(occ_final,cut_block==2),col=2,add=T)
plot(subset(occ_final,cut_block==3),col=3,add=T)
plot(subset(occ_final,cut_block==4),col=4,add=T)
Challenge: use your occurrences to cut raster layers
–load occurrences & raster layers
–build a600,000 meterbuffer around occurrences
–maskraster by the buffer of occurrences
–plot the masked rasterSolution
library(dismo) library(raster) dir.create("data") dir.create("data/bioclim") if(!file.exists("data/occ_raw.rdata")){ occ_raw <- gbif(genus="Dasypus",species="novemcinctus",download=TRUE) save(occ_raw,file = "data/occ_raw.rdata") }else{ load("data/occ_raw.rdata") } occ_clean <- subset(occ_raw,(!is.na(lat))&(!is.na(lon))) occ_unique <- occ_clean[!duplicated( occ_clean[c("lat","lon")] ),] occ_unique_specimen <- subset(occ_unique, basisOfRecord=="PRESERVED_SPECIMEN") occ_final <- subset(occ_unique_specimen, year>=1950 & year <=2000) coordinates(occ_final) <- ~ lon + lat myCRS1 <- CRS("+init=epsg:4326") # WGS 84 crs(occ_final) <- myCRS1 if( !file.exists( paste0("data/bioclim/bio_10m_bil.zip") )){ utils::download.file(url="http://biogeo.ucdavis.edu/data/climate/worldclim/1_4/grid/cur/bio_10m_bil.zip", destfile="data/bioclim/bio_10m_bil.zip" ) utils::unzip("data/bioclim/bio_10m_bil.zip",exdir="data/bioclim") } bio1 <- raster("data/bioclim/bio1.bil") occ_buffer <- buffer(occ_final,width=6*10^5) #unit is meter bio1_mask <- mask(bio1, occ_buffer) plot(bio1_mask) plot(occ_buffer,add=T) plot(occ_final,add=T,col="blue")